1 Pager settings
- Home
- Team
- Research
- Publications
- News
- Contact
- Contact
- Section name
- Section name
- Section name
- Section name
- Section name
- Section name
- Section name
- Section name
- Section name
- Section name
- Section name
- Section name
- Section name
- Section name
- Section name
- Section name
- Section name
- Section name
- Section name
- Section name
- Section name
- Section name
- Section name
NAZARKO LAB
Welcome to Autophagy Lab at Georgia State University!
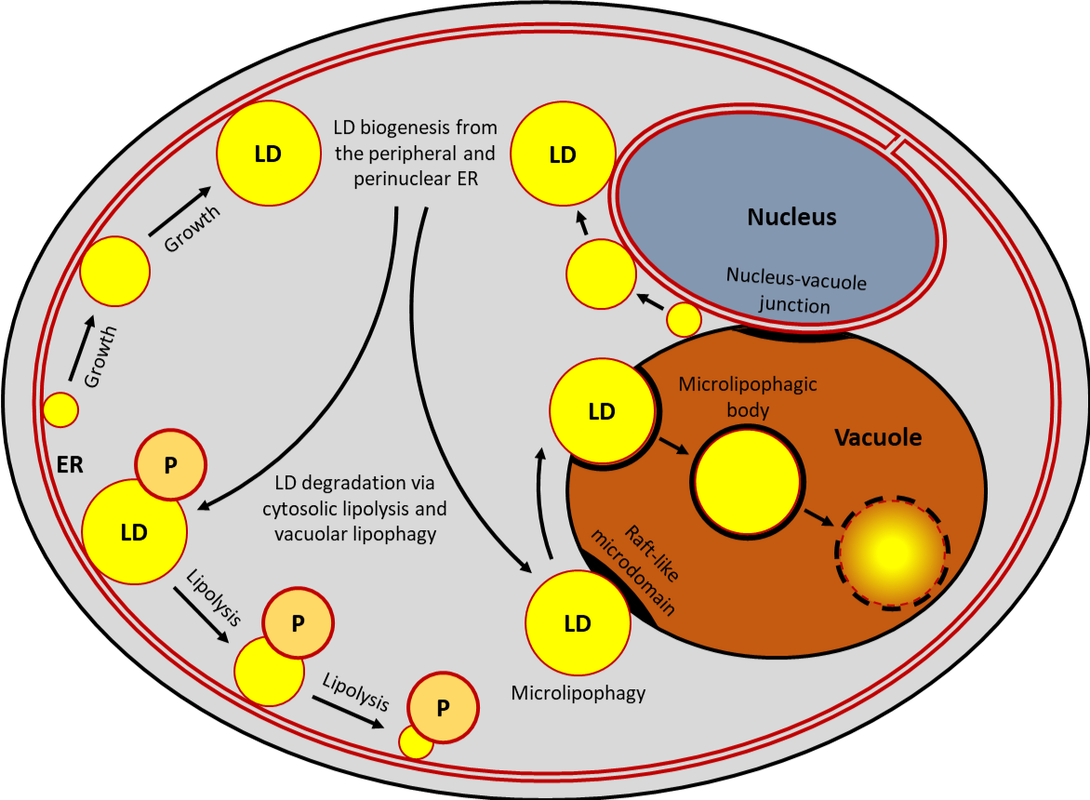
Our laboratory explores the selective autophagy of lipid droplets or lipophagy. This degradation pathway regulates lipid metabolism in most eukaryotic cells. Understanding molecular mechanisms of lipophagy is critical for controlling it in humans for the prevention and treatment of many lipid accumulation diseases, such as atherosclerosis, obesity and metabolic syndrome of aging.
Our laboratory explores the selective autophagy of lipid droplets or lipophagy. This degradation pathway regulates lipid metabolism in most eukaryotic cells. Understanding molecular mechanisms of lipophagy is critical for controlling it in humans for the prevention and treatment of many lipid accumulation diseases, such as atherosclerosis, obesity and metabolic syndrome of aging.